






 |
 |
|
 |
 |
 |
 |
 |
 |
CURRENT PROJECTS![]()
 1.
ETHANOL RESPONSIVE ELEMENTS IN THE MITOCHONDRIAL ASPARTATE AMINOTRANSFERASE
PROMOTER
1.
ETHANOL RESPONSIVE ELEMENTS IN THE MITOCHONDRIAL ASPARTATE AMINOTRANSFERASE
PROMOTER
When exposed to ethanol, hepatocytes increase their expression of mitochondrial
aspartate aminotransferase. This leads to an increase in the activity of this
enzyme in serum and is the biological basis for the characteristic rise in
AST seen in alcoholic liver disease. However, this enzyme has been found to
be identical to plasma membrane fatty acid binding protein, the first protein
identified as promoting facilitated uptake of long-chain free fatty acids
into cells. The increase in expression may also lead to hepatic steatosis
seen in alcoholic liver disease. This dual role makes this gene of extreme
interest in the study of alcoholic liver disease. Analysis of the promoter
is underway, to locate and define transcription factor binding sites that
may be involved in the response to ethanol. Reporter constructs with various
portions of the promoter regulating expression of secreted alkaline phosphatase
(SEAP) are employed, and transfected into human hepatoma cells. After selection,
cells are exposed to medium with 0 or 40 mM ethanol for 24 hours and medium
is assayed for SEAP activity using a fluorescent assay. Regions responsive
to ethanol will be analyzed for transcription factor binding sites using bioinformatics,
DNA Footprinting, EMSA, and various newer assay formats becoming available
to determine what factors alter promoter function. Comparisons may be made
with promoters of other genes known to be responsive to ethanol.
![]()
 2.
DELINEATION OF THE FATTY ACID BINDING SITE OF MITOCHONDRIAL ASPARTATE AMINOTRANSFERASE
2.
DELINEATION OF THE FATTY ACID BINDING SITE OF MITOCHONDRIAL ASPARTATE AMINOTRANSFERASE
Molecular modeling of mitochondrial aspartate aminotransferase has identified
a specific region in which a large number of hydrophobic residues face a cleft
in the surface. The volume of the cleft is suitable for binding a long-chain
fatty acid. Also, an arginine residue (R201) is present at one end, similar
to the placement of arginine or lysine residues in the fatty acid binding
sites of albumin. A rat cDNA clone has been mutated to alter specific residues
to their cognate forms found in the cytoplasmic isozyme, which has no significant
fatty acid binding capacity. Although the cytoplasmic and mitochondrial forms
catalyze the same reaction, they are only ~50% identical at the amino acid
sequence level. There are numerous residues that are conserved in either form,
where all mammals have a specific amino acid at one position in the mitochondrial
form, while all cytoplasmic forms share a different residue. In the cleft
region defined by just over 100 residues, 23 of these conserved substitutions
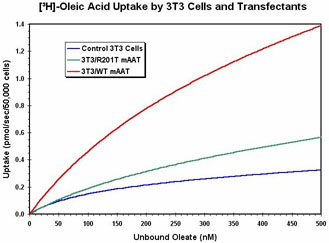
are present. Preliminary findings indicate that the substitutions R201T and
A219P decrease fatty acid binding and/or uptake substantially. A construct
with a complete replacement of the binding site region with cytoplasmic sequence
is also under investigation, as well as double mutants and a mutation, R201K,
which retains a basic residue at the key site. These structure-function studies
will help us determine how this single protein participates in two disparate
cellular processes.
![]()
 3.
HISTOLOGICAL ANALYSIS OF THE MOUSE GERM CELL LINEAGE
3.
HISTOLOGICAL ANALYSIS OF THE MOUSE GERM CELL LINEAGE
This project is based upon previous observations made while studying serial
sections of developing mouse fetuses and adult gonads. A by-product of a staining
reaction gave a specific staining pattern to mouse germ cells. This pattern
was seen in all stages from day 11 of fetal development onwards, and was present
in germ cells of all stages in adult gonads. These observations will be extended
to include the earlier stages of fetal and embryonic development, to determine
when it appears, or if it is maintained throughout development, which would
argue for a mode of continuity of the germ plasm. Also, another method employed
in paraffin sections appeared to delineate the acrosomal cap as it forms in
spermatogenesis, something not generally visible except in electron microscopy.
This method will be investigated for its compatibility with other staining
methods suitable for analysis spermatogenesis and may be useful in the study
of defects in that process.